vO.1.1 Release Notes¶
Introducing Blue Graph¶
This is an initial release of Blue Graph, a graph analytics framework for Python designed to unify different graph processing APIs. Its main goal is to provide a set of uniform APIs enabling users to peform graph analytics tasks abstracting away from the specificities of different graph processing tools, backends, and the graph data structures they provide.
Using the built-in PGFrame data structure for representing property graphs with node/edge types and property data types as a ‘data exchange layer’ between different analytics interfaces, it provides a backend-agnostic API supporting both in-memory (NetworkX, graph-tool and StellarGraph) and persistent graph backends (Neo4j).
It provides the following set of interfaces:
preprocessing and co-occurrence analysis API providing semantic property encoders and co-occurrence graph generators;
graph analytics API providing interfaces for computing graph metrics, performing path search and community detection;
unsupervised representation learning API for applying various graph embedding techniques;
representation learning downstream tasks API allowing the user to perform node classification, similarity queries, link prediction.
The main components of BlueGraph’s API are illustrated in the following diagram:
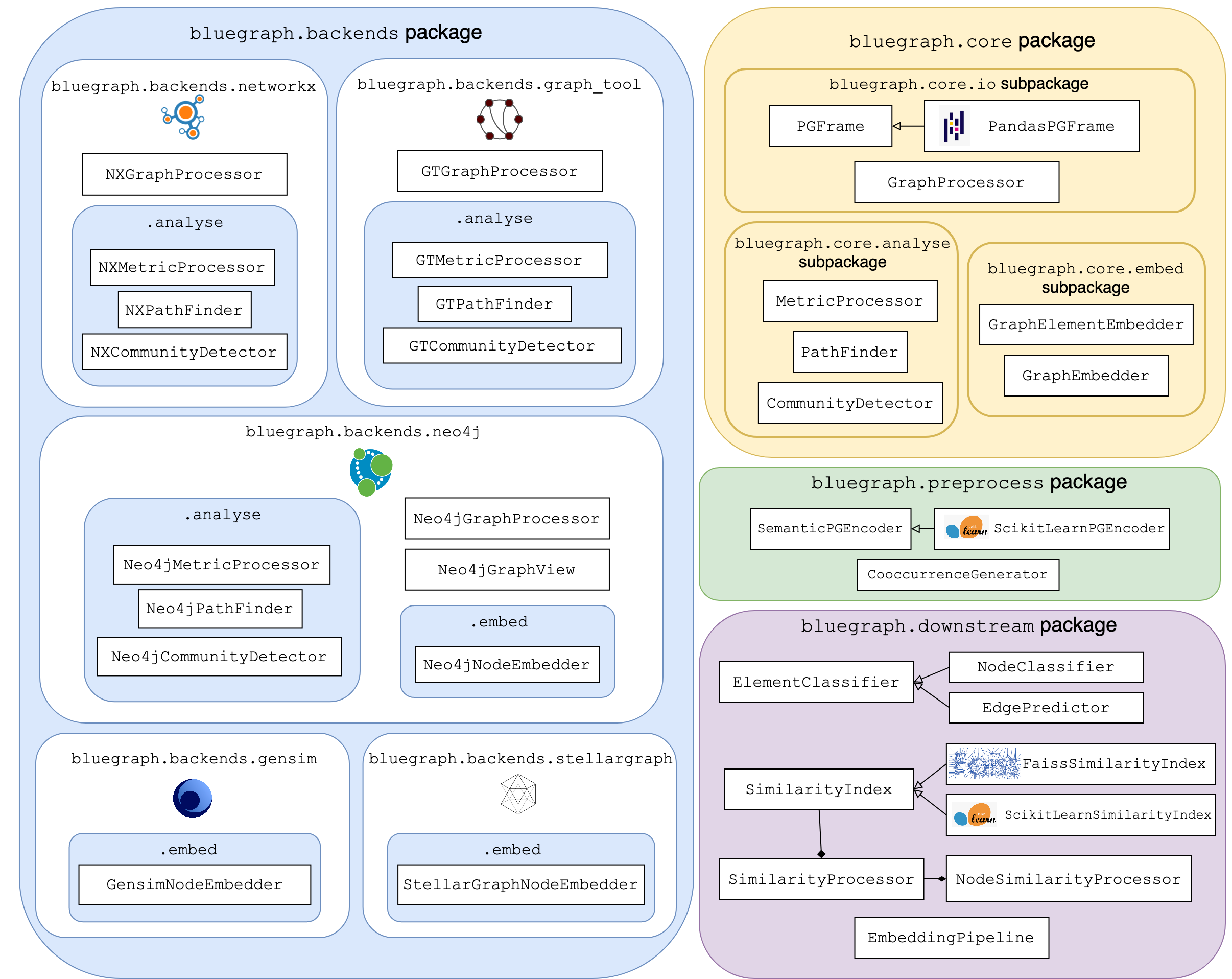
Blue Graph’s core¶
The bluegraph.core package provides the exchange graph data structure that serves as the input to graph processors based on different backends (PGFrame), as well as basic interfaces for different graph analytics and embedding classes (MetricProcessor, PathFinder, CommunityDetector, GraphElementEmbedder, etc).
PGFrame¶
This data structure represents property graphs with node/edge types and property data types. It is based on two dataframes: one for nodes, their types and properties as well as one for edges and their types/properties. Moreover, PGFrame supports setting data types to node/edge properties. Three high-level data types are currently supported: numeric for numerical properties (for example, age), category for properties representing categorical labels (for example, sex) and text for representing textual properties (for example, description).
This release includes a pandas-based implementation (PandasPGFrame) of the PGFrame data structure.
GraphProcessor¶
GraphProcessor is a basic interface for accessing and manipulating different graph elements (nodes/edges/properties) of backend-specific graph objects.
Graph preprocessing with BlueGraph¶
The bluegraph.preprocessing package provides a set of utils for preprocessing PGFrame graph objects.
Co-occurrence generation¶
The CooccurrenceGenerator interface allows the user to examine a provided property graph for co-occurrences (based on its nodes or edges), generate co-occurrence edges, and compute the following statistics:
co-occurrece factors,
co-occurrence frequency,
positive/normalized pointwise mutual information of the co-occurrence [4].
Semantic property encoding¶
The SemanticPGEncoder interface allows the user to convert properties of nodes and edges into numerical vectors. Currently, ScikitLearnPGEncoder is implemented in Blue Graph.
The encoder object can be instantiated as in the following example:
encoder = ScikitLearnPGEncoder(node_properties, edge_properties,
heterogeneous, drop_types,
encode_types, edge_features,
categorical_encoding,
text_encoding,
text_encoding_max_dimension,
missing_numeric,
imputation_strategy,
standardize_numeric)
Where the following parameters can be specified:
node_propertiesa list of node property names to encode;edge_propertiesa list of edge property names to encode;heterogeneousa flag indicating if the feature space is heterogeneous accross different node/edge types;encode_typesa flag indicating if node/edge types should be included in the generated features;categorical_encodingdefines a strategy for encoding categorical properties (currently, Multi Label Binarizer is available);text_encodingdefines a strategy for encoding text properties (currently, TfIdf and Doc2Vec encodings are available);missing_numerica strategy for treating missing numeric property values (dropping or imputing such values is possible);imputation_strategya strategy for imputing missing numeric property values (currently, only the mean imputation is available);standardize_numerica flag indicating if numerical property values should be standardized.
Backend support¶
Blue Graph supports the following in-memory and persistent graph analytics tools (see bluegraph.backends):
NetworkX (for the analytics API)
graph-tool (for the analytics API)
Neo4j (for the analytics and representation learning API);
StellarGraph (for the representation learning API).
NetworkX¶
The NetworkX-based graph analytics interfaces implemented in Blue Graph can be found in bluegraph.backends.networkx.
Conversion between PGFrame and NetworkX graph objects can be done using pgframe_to_networkx and networkx_to_pgframe. NXGraphProcessor allows accessing and manipulating graph elements of NetworkX graphs (bluegraph.backends.networkx.io).
Graph metrics¶
The NXMetricsProcessor interface supports the following graph metrics (all supporting unweighted and weighted graph edges):
graph density,
degree centrality,
PageRank centrality,
betweenness centrality,
closeness centrality.
Writing centrality measures as node properties is available.
Path search¶
The NXPathFinder interface supports the following path search tasks:
minimum spanning tree (unweighted and weighted, in-place labeling of the tree edges is available),
top neighbors by edge weight,
single shortest path (unweighted and weighted, excluding direct edge is available),
all shortest paths (excluding direct edge is available),
top shortest paths (unweighted and weighted, excluding direct edge is available, Naive [1] and Yen [3] strategies are available)
nested shortest paths [2] (unweighted and weighted, excluding direct edge is available, Naive and Yen strategies are available).
Community detection¶
The NXCommunityDetector interface supports the following community detection methods (all supporting both weighted and unweighted graphs):
Louvain algorithm (
strategy="louvain")Girvan–Newman algorithm (
strategy="girvan-newman")Label propagation (
strategy="lpa")Hierarchical clustering (
strategy="hierarchical", only for nodes with numerical features)
And supports the following partition quality metrics: modularity, performance, coverage.
Writing community labels as node properties is available.
graph-tool¶
The graph-tool-based analytics interfaces implemented in Blue Graph can be found in bluegraph.backends.graph_tool.
Conversion between PGFrame and graph-tool objects can be done using pgframe_to_graph_tool and graph_tool_to_pgframe. GTGraphProcessor allows accessing and manipulating graph elements of graph-tool graphs (bluegraph.backends.graph_tool.io).
Graph metrics¶
The GTMetricsProcessor interface supports the following graph metrics (all supporting unweighted and weighted graph edges):
graph density,
degree centrality,
PageRank centrality,
betweenness centrality,
closeness centrality.
Writing centrality measures as node properties is available.
Path search¶
The GTPathFinder interface supports the following path search tasks:
minimum spanning tree (unweighted and weighted, in-place labeling of the tree edges is available),
top neighbors by edge weight,
single shortest path (unweighted and weighted, excluding direct edge is available),
all shortest paths (excluding direct edge is available),
top shortest paths (unweighted and weighted, excluding direct edge is available, only the Naive [1] strategy is available)
nested shortest paths [2] (unweighted and weighted, excluding direct edge is available, the Naive strategy is available).
Community detection¶
The GTCommunityDetector interface supports the following community detection methods (supporting both weighted and unweighted graphs):
Statistical inference with Stochastic Block Models (
strategy="sbm") [5],Hierarchical clustering (
strategy="hierarchical", only for nodes with numerical features).
And supports the following partition quality metrics: modularity, performance, coverage.
Writing community labels as node properties is available.
Neo4j¶
The Neo4j-based graph analytics interfaces implemented in Blue Graph can be found in bluegraph.backends.neo4j and are based on the Neo4j Graph Data Science Library (version >= 1.5).
A Neo4j database instance can be populated from a PGFrame object using pgframe_to_neo4j and a PGFrame object can be read from a Neo4j database using neo4j_to_pgframe. Neo4jGraphProcessor allows accessing and manipulating graph elements of Neo4j graphs (bluegraph.backends.neo4j.io).
Graph metrics¶
The Neo4jMetricsProcessor interface supports the following graph metrics:
graph density (unweighted and weighted),
degree centrality (unweighted and weighted),
PageRank centrality (unweighted and weighted),
betweenness centrality (only unweighted),
closeness centrality (only unweighted).
Writing centrality measures as node properties is available.
Path search¶
The Neo4jPathFinder interface supports the following path search tasks:
minimum spanning tree (unweighted and weighted, only in-place labeling of the tree edges is available),
top neighbors by edge weight,
single shortest path (unweighted and weighted, excluding direct edge is available),
all shortest paths (excluding direct edge is available),
top shortest paths (unweighted and weighted, excluding direct edge is available, only the Yen [3] strategy is available)
nested shortest paths [2] (unweighted and weighted, excluding direct edge is available, only the Yen [3] strategy is available).
Community detection¶
The Neo4jCommunityDetector interface supports the following community detection methods (all supporting both weighted and unweighted graphs):
Louvain algorithm (
strategy="louvain")Girvan–Newman algorithm (
strategy="girvan-newman")Label propagation (
strategy="lpa")Hierarchical clustering (
strategy="hierarchical", only for nodes with numerical features)
And supports the following partition quality metrics: modularity, performance, coverage.
Writing community labels as node properties is available.
Node representation learning¶
The Neo4jNodeEmbedder interface supports the following unsupervised node representation models:
node2vec (transductive, only unweighted version),
FastRP (transductive),
GraphSAGE (inductive, model is saved in a model catalog of the current Neo4j instance).
StellarGraph¶
The StellarGraph-based graph representation learning interfaces implemented in Blue Graph can be found in bluegraph.backends.stellargraph.
Conversion between PGFrame and StellarGraph objects can be done using pgframe_to_stellargraph and stellargraph_to_pgframe (bluegraph.backends.stellargraph.io).
Node representation learning¶
The StellarGraphNodeEmbedder interface supports the following unsupervised node representation models:
node2vec (transductive, based on this demo),
Watch Your Step (transductive, based on this demo),
Deep Graph Infomax GCN, GAT, GraphSAGE (transductive, based on this demo),
attri2vec (inductive, based on this demo),
attri2vec (inductive, based on this demo),
GraphSAGE (inductive, based on this demo),
Deep Graph Infomax GCN & GAT with Cluster-GCN training procedure (inductive, based on this demo).
Downstream tasks with BlueGraph¶
Node classification API¶
The NodeClassifier interface provides a wrapper allowing to build classification models of PGFrame nodes.
The classifier object can be instantiated as in the following example:
classifier = NodeClassifier(model, feature_vector_prop, feature_props, **kwargs)
Where the following parameters can be specified:
modela classification model object that supports two methodsmodel.fit(data, labels)andmodel.fit(data)(for example, LinearSVC);feature_vector_propname of the feature vector property to use;feature_propsa list of property names (all must be numeric) to concatenate in order to obtain the node feature vectors.
Edge prediction API¶
The EdgePredictor interface provides a wrapper allowing for building edge prediction models for PGFrame graphs. Such models allow the user to discriminate between ‘true’ and ‘false’ edges based on embedding vectors of their source and target nodes.
The classifier object can be instantiated as in the following example:
classifier = EdgePredictor(model, feature_vector_prop, feature_props, operator, directed)
Where the following parameters can be specified:
modela classification model object that supports two methodsmodel.fit(data, labels)andmodel.fit(data)(for example, LinearSVC);feature_vector_propname of the node feature vector property to use;feature_propsa list of property names (all must be numeric) to concatenate in order to obtain the node feature vectors;operatorbinary operator to apply on the embedding vectors of source and target nodes (available operators, “hadamard”, “l1”, “l2” and “average”).
The generate_negative_edges util allows the user to generate ‘fake’ edges given a graph (that can be used for training an EdgePredictor).
Similarity API¶
The SimilarityProcessor interface allows building vector similarity indices using the Faiss Library. It wraps the indices (names or IDs) of the points, vector space and similarity measure configurations. It also allows segmenting the search space into Voronoi cells (see this example) allowing to speed up the search.
The NodeSimilarityProcessor is another wrapper that provides a higher-level abstraction to SimilarityProcessor and allows building and querying node similarity indices using Faiss. It wraps the underlying graph object and the vector similarity processor and provides an interface for querying similar nodes.
Building embedding pipelines¶
EmbeddingPipeline allows chaining the following steps and respective components:
property encoding (optional),
embedding model training (and prediction),
similarity index training (and querying).
Such embedding pipelines can also be saved and loaded.